Usage examples¶
Note
In the following examples, test_data can be replaced with the directory containing the FAST5 files
from your own runs. If you are new to ONT sequencing, the test_data directory is shipped with poretools
for experimentation.
poretools fastq¶
Extract sequences in FASTQ format from a set of FAST5 files.
poretools fastq test_data/*.fast5
Or, if there are too many files for your OS to do the wildcard expansion, just provide a directory.
poreutils will automatically find all of the FAST5 files in the directory.
poretools fastq test_data/
Extract sequences in FASTQ format from a set of FAST5 files.
poretools fastq test_data/
poretools fastq --min-length 5000 test_data/
poretools fastq --max-length 5000 test_data/
poretools fastq --type all test_data/
poretools fastq --type fwd test_data/
poretools fastq --type rev test_data/
poretools fastq --type 2D test_data/
poretools fastq --type fwd,rev test_data/
A type of “best” will extract the 2D read, if it exists. If not, it will extract either the template or complement read, whichever is available and has a better average Phred score.
poretools fastq --type best test_data/
Only extract sequence with more complement events than template. These are the so-called “high quality 2D reads” and are the most accurate sequences from a given run.
poretools fastq --type 2D --high-quality test_data/
The data in fastq format are returned in standard output.
poretools fasta¶
Extract sequences in FASTA format from a set of FAST5 files.
poretools fasta test_data/
poretools fasta --min-length 5000 test_data/
poretools fasta --max-length 5000 test_data/
poretools fasta --type all test_data/
poretools fasta --type fwd test_data/
poretools fasta --type rev test_data/
poretools fasta --type 2D test_data/
poretools fasta --type fwd,rev test_data/
poretools fasta --type best test_data/
The data in fasta format are returned in standard output.
poretools combine¶
Create a tarball from a set of FAST5 (HDF5) files.
# plain tar (recommended for speed)
poretools combine -o foo.fast5.tar test_data/*.fast5
# gzip
poretools combine -o foo.fast5.tar.gz test_data/*.fast5
# bzip2
poretools combine -o foo.fast5.tar.bz2 test_data/*.fast5
poretools yield_plot¶
Create a collector’s curve reflecting the sequencing yield over time for a set of reads. There are two types of plots. The first is the yield of reads over time:
poretools yield_plot --plot-type reads test_data/
The result should look something like:

The second is the yield of base pairs over time:
poretools yield_plot --plot-type basepairs test_data/
The result should look something like:
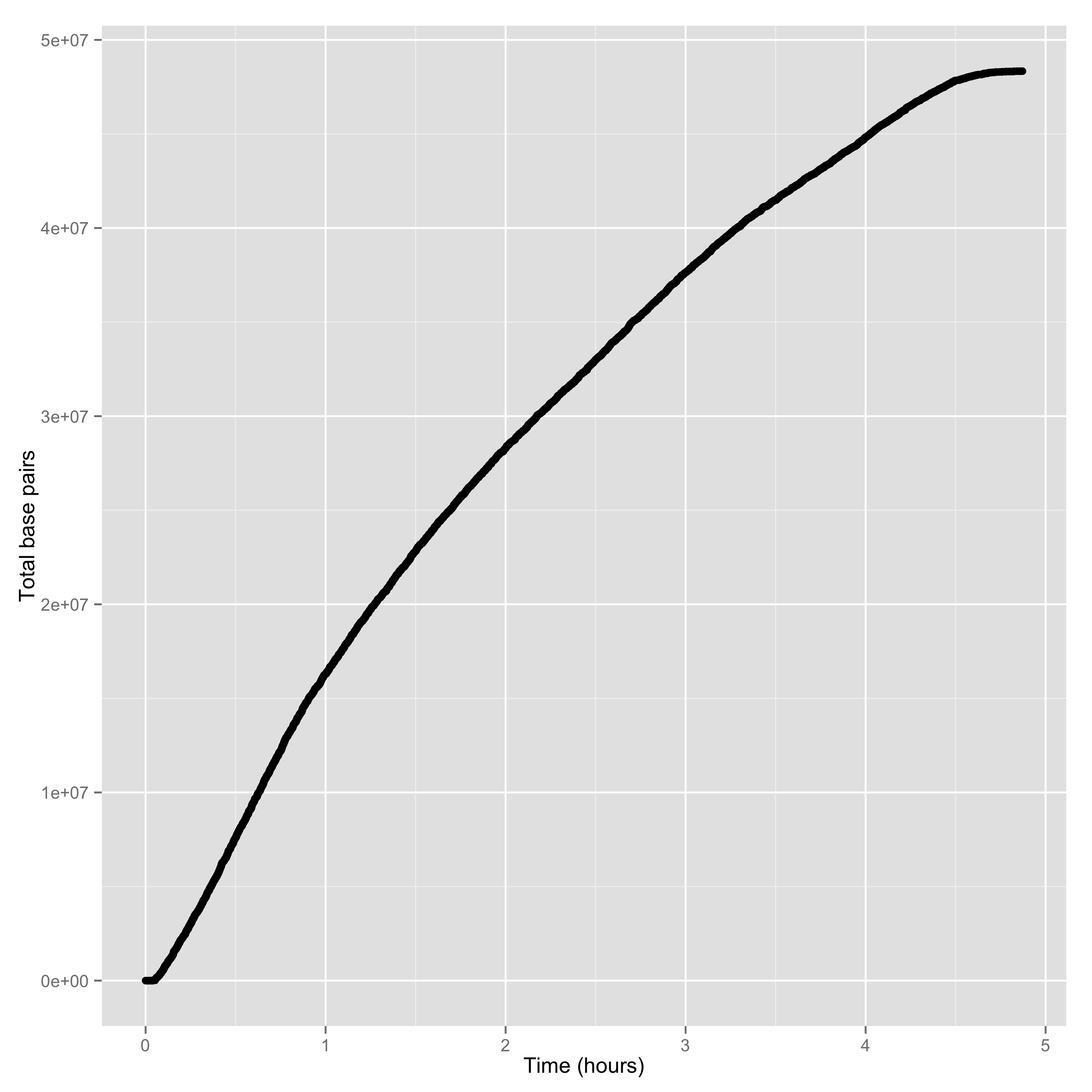
Of course, you can save to PDF or PNG with –saveas:
poretools yield_plot \
--plot-type basepairs \
--saveas foo.pdf\
test_data/
poretools yield_plot \
--plot-type basepairs \
--saveas foo.png\
test_data/
If you don’t like the default aesthetics, try –theme-bw:
poretools yield_plot --theme-bw test_data/
poretools squiggle¶
Make a “squiggle” plot of the signal over time for a given read or set of reads
poretools squiggle test_data/foo.fast5
The result should look something like:
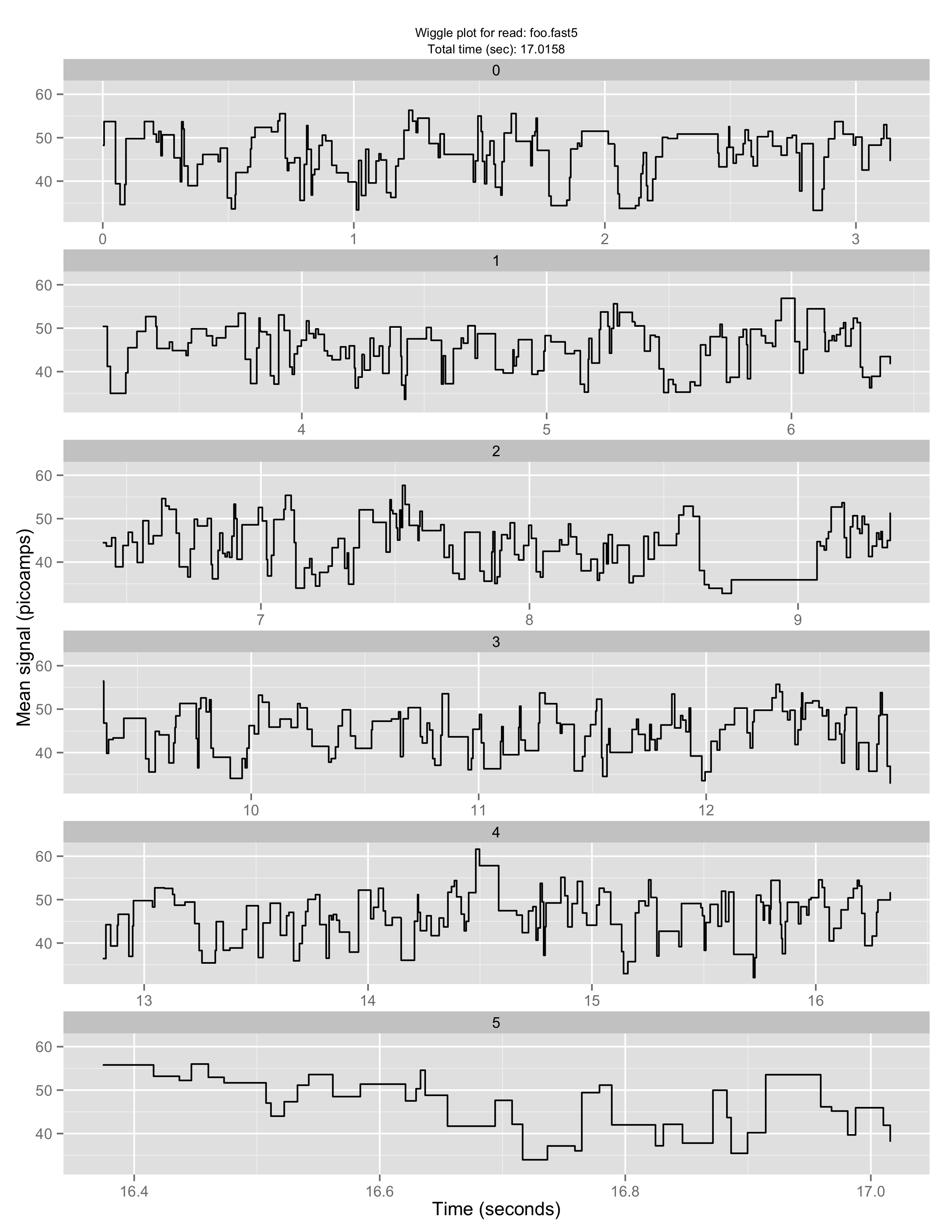
If you don’t like the default aesthetics, try –theme-bw:
poretools squiggle --theme-bw test_data/
Other options:
# save as PNG
poretools squiggle --saveas png test_data/foo.fast5
# save as PDF
poretools squiggle --saveas pdf test_data/foo.fast5
# make a PNG for each FAST5 file in a directory
poretools squiggle --saveas png test_data/
poretools winner¶
Report the longest read among a set of FAST5 files.
poretools winner test_data/
poretools winner --type all test_data/
poretools winner --type fwd test_data/
poretools winner --type rev test_data/
poretools winner --type 2D test_data/
poretools winner --type fwd,rev test_data/
poretools winner --type best test_data/
poretools stats¶
Collect read size statistics from a set of FAST5 files.
poretools stats test_data/
total reads 2286.000000
total base pairs 8983574.000000
mean 3929.822397
median 4011.500000
min 13.000000
max 6864.000000
poretools hist¶
Plot a histogram of read sizes from a set of FAST5 files.
poretools hist test_data/
poretools hist --min-length 1000 --max-length 10000 test_data/
poretools hist --num-bins 20 --max-length 10000 test_data/
If you don’t like the default aesthetics, try –theme-bw:
poretools hist --theme-bw test_data/
The result should look something like:

poretools nucdist¶
Look at the nucleotide composition of a set of FAST5 files.
poretools nucdist test_data/
A 78287 335291 0.233489714904
C 75270 335291 0.224491561062
T 92575 335291 0.276103444471
G 84754 335291 0.252777438106
N 4405 335291 0.0131378414571
poretools qualdist¶
Look at the quality score composition of a set of FAST5 files.
poretools qualdist test_data/
! 0 83403 335291 0.248748102395
" 1 46151 335291 0.137644613187
# 2 47463 335291 0.141557632027
$ 3 34471 335291 0.102809201559
% 4 24879 335291 0.0742012162569
& 5 20454 335291 0.0610037251224
' 6 16783 335291 0.0500550268274
( 7 13699 335291 0.0408570465655
) 8 11356 335291 0.0338690868529
* 9 9077 335291 0.0270720061081
+ 10 6492 335291 0.0193622852984
, 11 4891 335291 0.014587328619
- 12 3643 335291 0.0108651887465
. 13 2585 335291 0.00770972080968
/ 14 1969 335291 0.0058725107444
0 15 1475 335291 0.00439916371152
1 16 1146 335291 0.00341792651756
2 17 902 335291 0.00269020045274
3 18 790 335291 0.00235616225905
4 19 619 335291 0.0018461575169
5 20 532 335291 0.00158668142002
6 21 440 335291 0.00131229290378
7 22 397 335291 0.00118404609727
8 23 379 335291 0.00113036138757
9 24 313 335291 0.000933517452004
: 25 327 335291 0.000975272226215
; 26 138 335291 0.000411582774366
< 27 121 335291 0.000360880548538
= 28 96 335291 0.000286318451733
> 29 76 335291 0.000226668774289
? 30 69 335291 0.000205791387183
@ 31 61 335291 0.000181931516205
A 32 48 335291 0.000143159225866
B 33 23 335291 6.8597129061e-05
C 34 14 335291 4.17547742111e-05
D 35 6 335291 1.78949032333e-05
F 37 3 335291 8.94745161666e-06
poretools qualpos¶
Produce a box-whisker plot of qualoty score distribution over positions in reads.
poretools qualpos test_data/
The result should look something like:
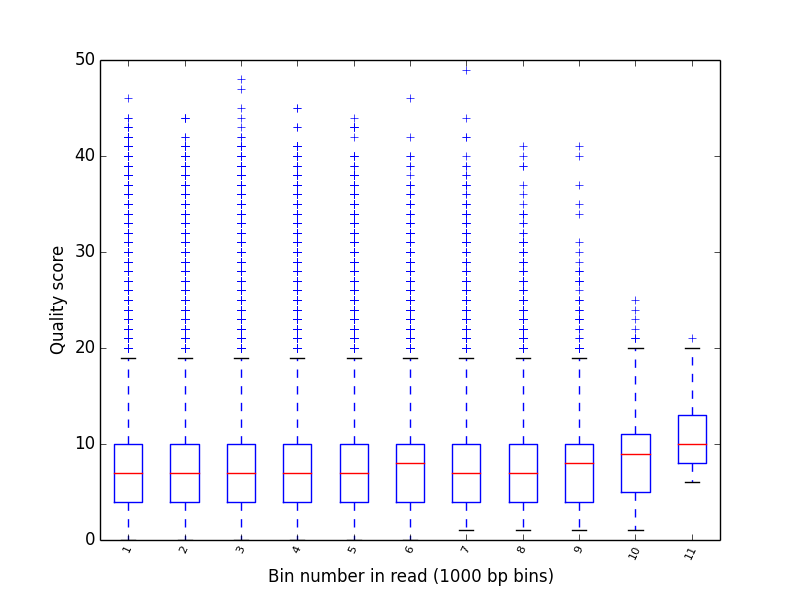
poretools tabular¶
Dump the length, name, seq, and qual of the sequence in one or a set of FAST5 files.
poretools tabular foo.fast5
length name sequence quals
10 @channel_100_read_14_complement GTCCCCAACAACAC $%%'"$"%!)
poretools events¶
Extract the raw nanopore events from each FAST5 file.
poretools events test_data/ | head -5
file strand mean start stdv length model_state model_level move p_model_state mp_model_state p_mp_model_state p_A p_C p_G p_T raw_index
test_data/2016_3_4_3507_1_ch120_read240_strand.fast5 template 58.3245290305 1559.89409031 1.34165996292 0.0146082337317 CGACTT 58.1304809188 0 0.0226559 CATCTT 0.0229866 0.284469 0.130683 0.137386 0.447461
test_data/2016_3_4_3507_1_ch120_read240_strand.fast5 template 50.1420877511 1559.90869854 0.921372775302 0.0348605577689 GACTTT 49.3934875964 1 0.0849836 GACTTT 0.0849836 0.257314 0.350541 0.101351 0.290794
test_data/2016_3_4_3507_1_ch120_read240_strand.fast5 template 47.5841029424 1559.9435591 0.771398562801 0.00763612217795 ACTTTG 48.2080162623 1 0.108899 TCTTTG 0.13079 0.000477931 0.00853333 0.306356 0.684632
test_data/2016_3_4_3507_1_ch120_read240_strand.fast5 template 51.5879264562 1559.95119522 0.684238307171 0.0112881806109 CTTTGA 52.7784154546 1 0.110625 CTTTGG 0.121103 4.69995e-06 0.00382846 0.0169048 0.979262
Extract the pre-basecalled events from each FAST5 file.
poretools events --pre-basecalled test_data/ | head -5
file strand mean start stdv length model_state model_level move p_model_state mp_model_state p_mp_model_state p_A p_C p_G p_T raw_index
burn-in-run-2/ch100_file15_strand.fast5 pre_basecalled 51.4652695313 5352344 0.655003995591 35
burn-in-run-2/ch100_file15_strand.fast5 pre_basecalled 60.1776123047 5352379 1.05143911309 18
burn-in-run-2/ch100_file15_strand.fast5 pre_basecalled 48.9152374359 5352397 0.864834628834 67
burn-in-run-2/ch100_file15_strand.fast5 pre_basecalled 55.4002178596 5352464 1.75915620083 17
poretools times¶
poretools times test_data/ | head -5
channel filename read_length exp_starttime unix_timestamp duration unix_timestamp_end iso_timestamp day hour minute
120 test_data/2016_3_4_3507_1_ch120_read240_strand.fast5 5826 1457127309 1457128868 47 1457128915 2016-03-04T15:01:08-0700 04 15 01
120 test_data/2016_3_4_3507_1_ch120_read353_strand.fast5 3399 1457127309 1457129863 28 1457129891 2016-03-04T15:17:43-0700 04 15 17
120 test_data/2016_3_4_3507_1_ch120_read415_strand.fast5 2640 1457127309 1457130808 24 1457130832 2016-03-04T15:33:28-0700 04 15 33
120 test_data/2016_3_4_3507_1_ch120_read418_strand.fast5 3487 1457127309 1457130851 31 1457130882 2016-03-04T15:34:11-0700 04 15 34
poretools occupancy¶
Plot the throughput performance of each pore on the flowcell during a given sequencing run.
poretools occupancy test_data/
The result should look something like:
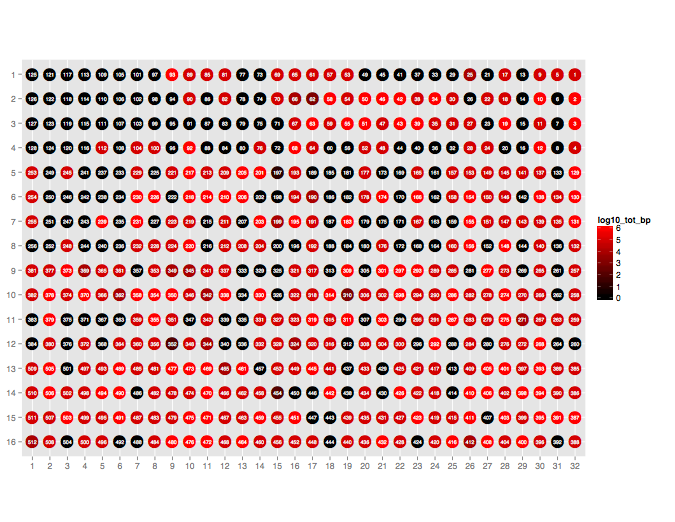
poretools index¶
Tabulate all file location info and metadata such as ASIC ID and temperature from a set of FAST5 files
poretools index test_data | head -5 | column -t
source_filename template_fwd_length complement_rev_length 2d_length asic_id asic_temp heatsink_temp channel exp_start_time exp_start_time_string_date exp_start_time_string_time start_time start_time_string_date start_time_string_time duration fast5_version
test_data/2016_3_4_3507_1_ch120_read240_strand.fast5 5826 5011 5079 3571011476 30.37 36.99 120 1457127309 2016-Mar-04 (Fri) 14:35:09 1457128868 2016-Mar-04 (Fri) 15:01:08 47 metrichor1.16
test_data/2016_3_4_3507_1_ch120_read353_strand.fast5 3399 2962 2940 3571011476 30.37 36.99 120 1457127309 2016-Mar-04 (Fri) 14:35:09 1457129863 2016-Mar-04 (Fri) 15:17:43 28 metrichor1.16
test_data/2016_3_4_3507_1_ch120_read415_strand.fast5 2640 2244 2428 3571011476 30.37 36.99 120 1457127309 2016-Mar-04 (Fri) 14:35:09 1457130808 2016-Mar-04 (Fri) 15:33:28 24 metrichor1.16
test_data/2016_3_4_3507_1_ch120_read418_strand.fast5 3487 2950 3384 3571011476 30.37 36.99 120 1457127309 2016-Mar-04 (Fri) 14:35:09 1457130851 2016-Mar-04 (Fri) 15:34:11 31 metrichor1.16
Extract the metadata from the fast5 file
poretools metadata 013731_11rx_v2_3135_1_ch20_file19_strand.fast5
asic_id asic_temp heatsink_temp
31037 28.11 37.88
poretools metadata --read 013731_11rx_v2_3135_1_ch20_file19_strand.fast5
filename scaling_used abasic_peak_height hairpin_polyt_level median_before start_time read_id read_number hairpin_peak_height abasic_found abasic_event_index duration start_mux hairpin_found hairpin_event_index
013731_11rx_v2_3135_1_ch20_file19_strand.fast5 1 124.31769966 0.413218809334 226.393825112 4648221 3b4e45bf-6d42-45bc-9314-1d8a630971c2 19 125.783167256 1 2 195322 4 1 1478